A Precisely Controlled DNA Biped
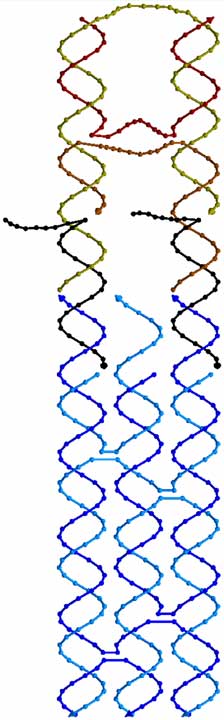 Shown
at left is a 3D drawing of the phosphate backbone of the first DNA biped. Each
base is represented by a sphere along the backbone (or a cone for the base on
the 3’ end of each strand). The biped walks along a footpath, shown in shades
of blue. In this case, the footpath is a TX molecule. The biped, shown in tan
and brown strands, has two double helical domains connected to each other by 3
flexible linkers made out of single strands of DNA (strands which have not
mated up to form double helices). At the bottom of the biped are two single
stranded sections which are complementary to black “set” strands. The set
strands, in turn, are complementary to single stranded regions on the top of
the footpath. By selecting the correct set strands, one can control which foot
of the biped is connected to which foothold on the footpath.
Shown
at left is a 3D drawing of the phosphate backbone of the first DNA biped. Each
base is represented by a sphere along the backbone (or a cone for the base on
the 3’ end of each strand). The biped walks along a footpath, shown in shades
of blue. In this case, the footpath is a TX molecule. The biped, shown in tan
and brown strands, has two double helical domains connected to each other by 3
flexible linkers made out of single strands of DNA (strands which have not
mated up to form double helices). At the bottom of the biped are two single
stranded sections which are complementary to black “set” strands. The set
strands, in turn, are complementary to single stranded regions on the top of
the footpath. By selecting the correct set strands, one can control which foot
of the biped is connected to which foothold on the footpath.
Any given set strand can be pulled from the system by introducing into the system an “unset” strand. The set strand with the appropriate base sequence binds to the unset strand more strongly than to the biped/footpath, so it gets pulled out of the system. This process allows the operator to selectively pick up one foot or the other. A detached foot can then be placed on an available foothold by the introduction of the appropriate set strand.
A cartoon of the bipped taking a step is illustrated below. Here, the different sequences of the two feet of the biped are represented by pink and orange, while the different sequences of the 3 footholds are represented by different shades of blue/green. Complementary sequences are shown having the same color. The biped starts with foot 1 on foothold A, and foot 2 in foothold B. This is called state 1A,2B. In panel b, the unset strand has been introduced to remove foot 2 from foothold B. First, the unset strand grabs the unmated bases in set strand 2B (shown as a gray square). Then, as shown in panel c, the unset strand completely binds to the set strand, removing it from the complex. The bound set/unset strands can then be removed from the solution via the use of magnetic streptavidin beads, which bind the biotins on the ends of the unset strands (shown as blue circles). Once foot 2 is detached, it can be placed on foothold C, which brings the biped to state 1A,2C, as shown in panel d (this is also the state illustrated in the 3D drawing at left). When unset strand 1A is introduced into the system, foot 1 is detached from foothold A as shown in panel e. Then foot 1 can be attached to foothold B, as shown in panel f. Over the course of this step, the biped travels approximately 2 nanometers. Because the rise and fall of each foot is precisely controlled by the specific base sequences of the set and unset strands, the step is completely reversible, and the biped can walk back to where it started.
Each foot of the biped has a psoralen on the bottom. When exposed to UV light, psoralen will covalently link the foot to the foothold it stands upon. The state of the biped can then easily be monitored using chromatography to separate out the largest covalently linked structures. This work is in the July 2004 issue of Nano Letters.
For a news video about the biped, including a nice movie showing the action of the biped as it walks, see ScienceCentral News.
In the future, we hope to build larger systems with multiple
bipeds walking on larger footpaths. Ultimately these devices may be used to
move molecules around for precise nano-scale construction and 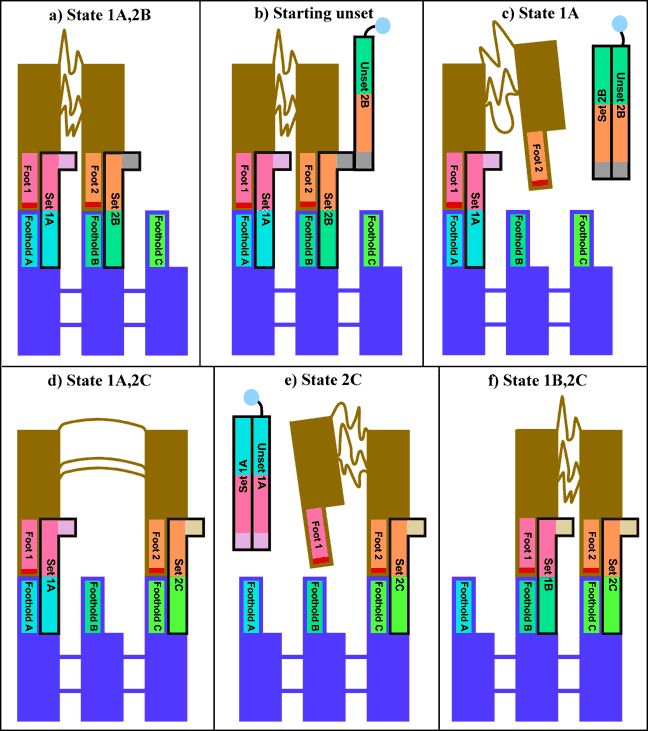 manufacturing.
manufacturing.