A DNA Nanomechanical Device
Based on Hybridization Topology
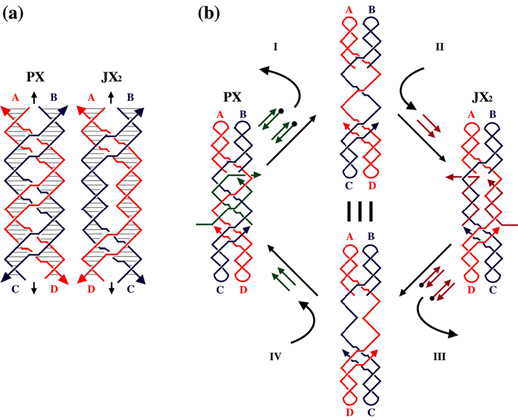
The sequence-dependent device is based on the PX motif of DNA. The PX motif, postulated to be involved in genetic recombination, consists of two helical domains formed by four strands that flank a central dyad axis (indicated by the vertical black arrows). In (a) below, two stands are drawn in red and two in blue, where the arrowheads indicate the 3' ends of the strands. The Watson-Crick base pairing in which every nucleotide participates is indicated by the thin horizontal lines within the two double helical domains. Every possible crossover occurs between the two helical domains. The same conventions apply to the JX2 motif which lacks two crossovers in the middle. The letters A, B, C and D, along with the color coding, show that the bottom of the JX2 motif (C and D) are rotated 180û relative to the PX motif. (b) illustrates the principles of device operation. On the left is a PX molecule. The green set strands are removed by the addition of biotinylated green fuel strands (biotin indicated by black circles) in process I. The unstructured intermediate is converted to the JX2 motif by the addition of the purple set strands in process II. The JX2 molecule is converted to the unstructured intermediate by the addition of biotinylated purple fuel strands in process III. The identity of this intermediate and the one above it is indicated by the identity sign between them. The cycle is completed by the addition of green set strands in process IV, restoring the PX device.
Return to
Ned Seeman's Nanotechnology Page
Return to
Homepage for Ned Seeman's Lab

