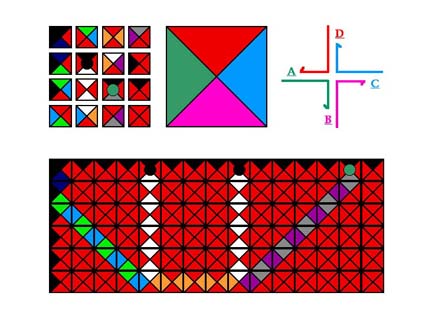
Shown below in the upper left are a group of Wang tiles based on a diagram in "Tilings and Patterns" by Grunbaum and Shephard. The edges of these tiles are colored, sometimes with a single color, and sometimes with as many as four different colors. The tiles can form a mosaic, such as the one shown at the bottom, according to the rule that each edge in the mosaic is flanked by the same color. This type of assembly can be shown to emulate the operation of a Turing machine. The mosaic shown is a simple addition. If one counts columns, one can see that there is a special tile in the top row in the 5th and 9th column, and a somewhat different special tile in the top row of the 14th column, corresponding to the sum of 5 + 9. If the tiles associate to form correct associations according to the rule, the rightmost special tile will occur at the position corresponding to the sum of the other two tiles. To do precisely the sum of 5 + 9, and not, say, 4 + 8, the 2nd through 8th tiles in the top row would all need to be unique. Nevertheless, about 23 tiles could result in the formation of a pattern containing 84 tiles if tiles associated in this way. Several years ago, Erik Winfree pointed out that our system, branched DNA with sticky ends, might lead to reducing this theoretical construct to practice. Note that the colors of the sticky ends on the branched junction in the upper right are the same as those on the large Wang tile in the center.

Recently, a version of this type of computation was reduced to practice in this laboratory. We performed a cumulative XOR calcaulation. In a cumulative XOR, the current value of Y, Y(i) = XOR [Y(i-1),X(i)]. The XOR operation reports a 1 if the two inputs are different, 0 and 1 or 1 and 0, and it reports a 0 if they are the same, 1 and 1 or 0 and 0. The calculation was performed with the components below.
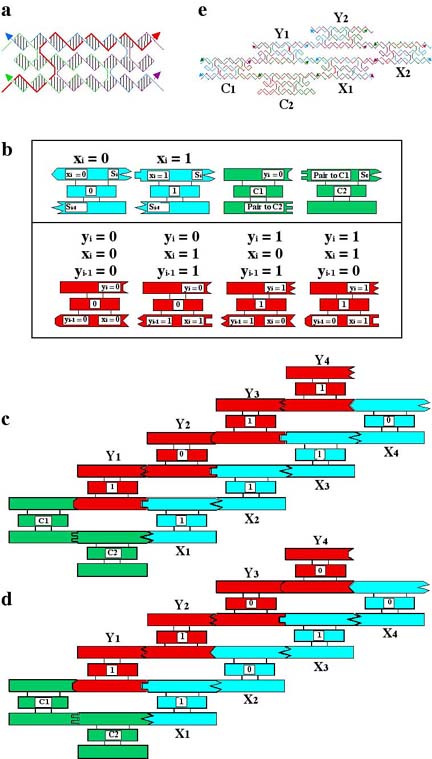
(a) shows a triple crossover (TX) molecule, whose topology produces a reporter strand that is drawn with a thick red line. Each of the components is a similar triple crossover molecule. (b) shows the various TX molecules in schematic. The input X(i) tiles are drawn in light blue. Their value is shown at the center of the tile. A '1' means chemically that the reporter strand in that tile contains a restriction site for EcoR V and a '0' tile contains a site for Pvu II. Each X tile in this calculation was specifically targetted for its position, by giving it individual sticky ends. To perform the calculation in parallel, one would allow the X(i) tiles to associate randomly, giving, for a four-bit problem, 16 different final answer strands. The X-tiles are connected to the Y tiles by the two green connector tiles to their right. The four possible Y answer tiles, corresponding to the two different inputs are drawn below these tiles. Note that both sticky ends must count in determining the Y tile that goes into the correct slot. The sticky end (drawn as a geometrical shape) that corresponds to, say, X(i) = 1 is the same both for the second tile [Y(i-1) = 1; X(i) = 1; -->Y(i) = 0] and for the fourth tile [Y(i-1) = 0; X(i) = 1; --> Y(i) = 1]. To ensure that the impact of both sticky ends was felt, longer sticky ends were assigned to those tiles connecting the X and C tiles, than to those involving the Y tiles; hence, the X and C tiles assembled first in a cooling protocol.
The two calculations we performed are shown in (c) and (d) above. In (c), the inputs are X(1) = 1; X(2) = 1; X(3) =1; X(4) = 0. This should lead to Y(1) = 1; Y(2) = 0; Y(3) = 1; Y(4) = 1. In (d), the input values are X(1) = 1; X(2) = 0; X(3) = 1; X(4) = 0, leading to Y(1) = 1; Y(2) = 1, Y(3) = 0; Y(4) = 0.
To extract the answer from the self-assembly, some means must be found to make a record of it. This is done by ligating the reporter strands together, as shown at the corner in (e). Once the ligation is complete, the strand is subjected to PCR and is treated with the appropriate restriction enzymes. The answers to the two calculations are then visible on a gel, as shown below:
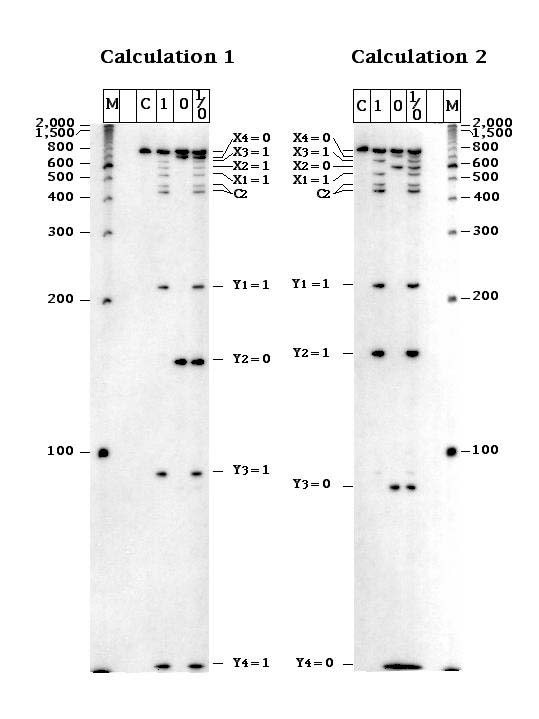
Thus, it is possible to do computation by self-assembly of DNA tiles. The success of this experiment indicates that it is apossible to use DNA components to do algorithmic assembly, in addition to periodic assembly.
Go to Ned Seeman's Nanotechnology Page
Return to Homepage for Ned Seeman's Lab